


Scientists have found that various signaling pathways can precisely regulate embryonic development and organ formation in time and space. In recent years, CRISPR related gene editing technology provides a good platform for the study of human development.
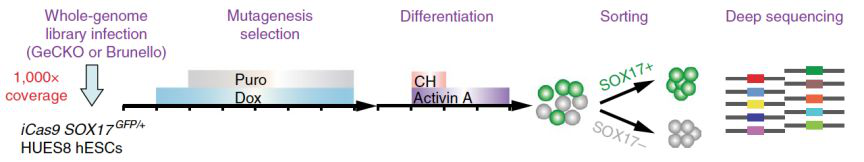
The researchers designed a library of more than 76000 gRNAs and transferred them into ESC to target every gene in the whole genome. After activating gene editing, they used activin A and chir99021 (GSK3 inhibitors) to induce ESC to differentiate into De. They then used FACS to separate differentiated and undifferentiated cells based on the expression of SOX17 gene. By comparing the proportion of various gRNAs in the two kinds of cells, the researchers found 37 positive regulatory factors (promoting differentiation) and 28 reverse regulatory factors (preventing differentiation), and confirmed most of them in turn.
Among the 6 reverse regulatory factors with FDR < 0.05, 4 are in jnk-jun pathway, including MEKK1, MKK4, MKK7 and Jun. As a verification, the researchers inactivated jnk-jun pathway through gene knockout and drug inhibition, which directly led to the differentiation rate of de increased to more than 90%.
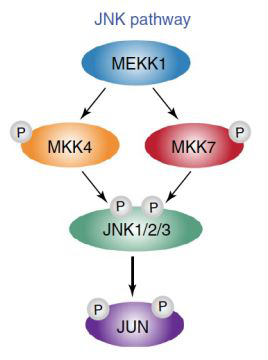
Finally, the jnk-jun pathway regulating de differentiation was screened by CRISPR / CAS technology, and it was explained that jnk-jun pathway was regulated by combining ESC promoter. This work also reveals the value of CRISPR genome-wide screening in developmental biology.
Case 1: the role of CRISPR genomic screening signaling pathway research
Case 2: the role of CRISPR genome screening in the discovery of new tumor immune regulatory factors